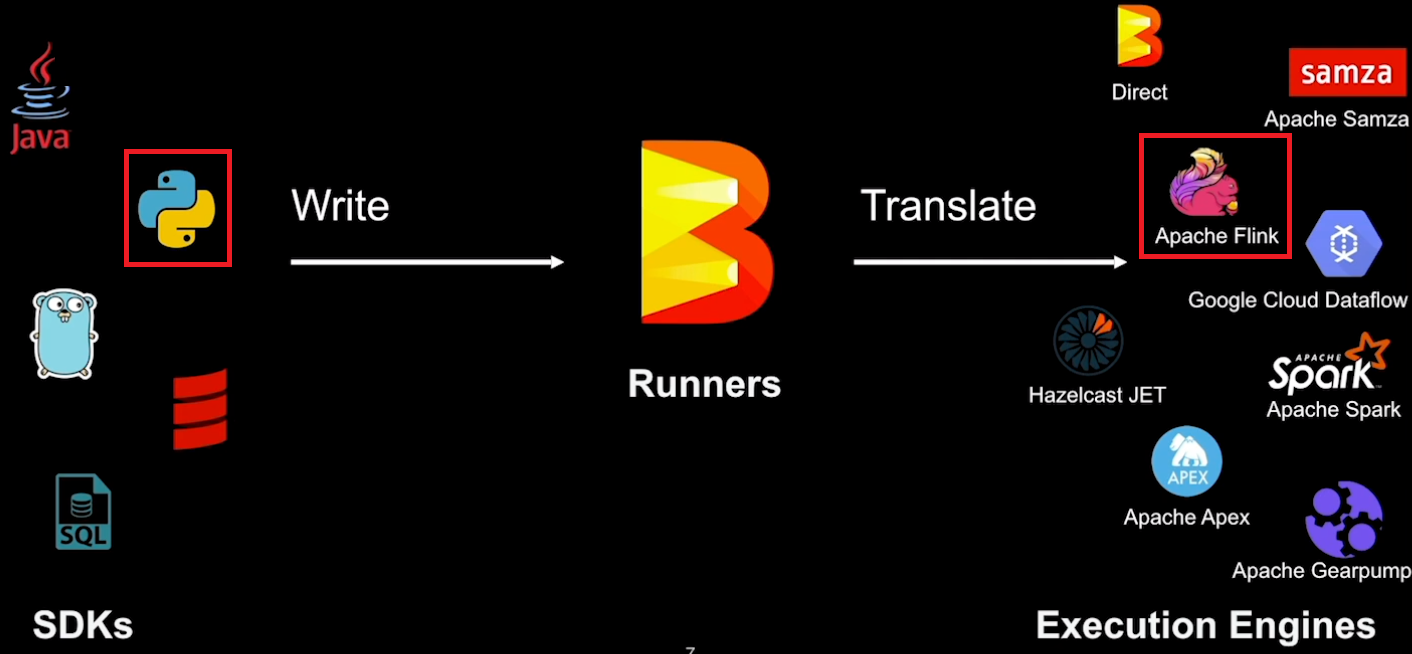
In the previous post, we developed an Apache Beam pipeline where the input data is augmented by a Remote Procedure Call (RPC) service. Each input element performs an RPC call and the output is enriched by the response. This is not an efficient way of accessing an external service provided that the service can accept more than one element. In this post, we discuss how to enhance the pipeline so that a single RPC call is made for a bundle of elements, which can save a significant amount time compared to making a call for each element.